Earlier this year, Frost and Sullivan predicted the global digital pathology market will grow from $513 million in 2019 to $826 million by 2025. Largely fuelled by the drive to boost lab efficiencies and an increasing prevalence of cancer, this 8.2 per cent compound annual growth rate lies on the conservative side of other forecasts, which point to double-digit growth rates and market sizes that reach $1.3 billion by 2028.
Double-digit growth or not, these figures spell good news for manufacturers of hardware, such as whole slide imagers, as well as associated software and storage systems. At the same time, the burgeoning market is also sparking the development of novel devices set to ease life in the lab. A quick look at recent products supports the rosy outlook.
In August 2020, Leica Biosystems launched its Aperio GT 450 DX digital pathology scanner, which aims to increase throughput, reduce turnaround times and generate high-quality images in primary diagnosis. Less than six months later, Roche delivered two artificial intelligence-based uPath image analysis algorithms that, combined with the company’s Ventana DP 200 slide scanner and uPath enterprise software, are set to deliver precision patient diagnosis in breast cancer. Roche also introduced Digital Pathology Open Environment so software developers can integrate image analysis tools for tumour tissue with its enterprise software.
‘We’ve been at the early stages of digital pathology for a while now,’ said Mike Rivers, vice president and lifecycle leader, digital pathology, from Roche’s Ventana Medical Systems. ‘The market has been held back by factors such as IT infrastructure and storage costs, but these [issues] are resolving in the right direction, the technology is maturing and people are really seeing the opportunities.
‘Ultimately we want to enable pathologists to take advantage of the digital environment and do things that they can’t do manually... This could be synchronising multiple digital images together and allowing them to make annotations on multiple sequential sections of tissue with a single annotation,’ he added.
Ensuring that whole slide imaging and software can be integrated into a clinical setting’s existing pathology workflow is critical, as is the move towards automated image analysis, to reduce turnaround times. And along the way, artificial intelligence will play a key role.
‘Scanners have already improved dramatically in terms of speed and image quality so AI is going to be the next big thing that will push us past the tipping point – we have a good example of image analysis algorithms making a difference in breast pathology,’ Rivers said.
Indeed, in October this year, Roche joined forces with pathology AI developers, PathAI and Ibex Medical Analytics, to develop embedded image analysis workflows that can be accessed via the cloud version of its uPath enterprise software, Navify Digital Pathology. Rivers is convinced AI algorithms will particularly assist pathologists in quantifying data.
‘In a tumour micro-environment you often want to see multiple biomarkers at the same time... This can become difficult for the human eye to distinguish, but effective use of image analysis and AI to, say, convolute the colours, provide proximity analysis and other measurements of interest, could ultimately be very powerful,’ he said.
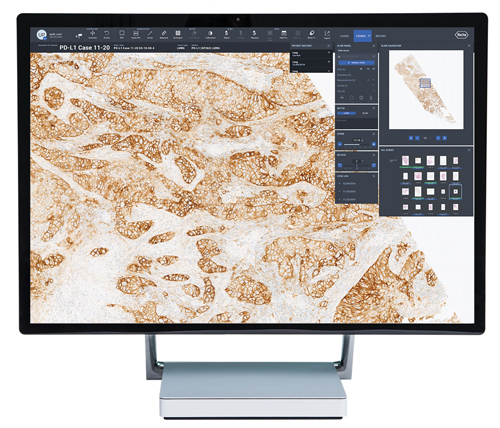
Viewing lung tissue using uPath software from Roche. Credit: Roche
As part of a digital pathology workflow, AI could also be used to prioritise a pathologist’s caseload and allow an algorithm to perform the tedious task of counting cell division, or mitoses. Still, uncertainties exist around AI and the jury is also out on when fully digital workflows will become mainstream. A few labs are leading the way but widespread adoption is probably several years away. As Rivers said: ‘We’ll see significant movement in the next five years and, in the labs that are already somewhat automated and are willing to invest, we’re going to see a massive transition in the next decade.’
Beyond pathology
But as pathologists around the world get ready for the inevitable digital overhaul, many tech businesses are also developing new imaging-related devices and systems that promise to reduce analysis times in the lab. A case in point is technology consultancy, Cambridge Consultants, which has developed PureSentry, a contamination detection system for cell and gene therapy monitoring that uses an event-based sensor from French firm Prophesee.
As Cambridge Consultants’ senior physicist, Josh Gibson, pointed out, he and colleagues wanted to develop a system that would cut the time and costs of arduous sterility testing in this sector. This process firstly demands a ten-day culture period followed by the actual sterility test, which can also take up to another ten days.
Gibson said: ‘In the gene therapy market, each dose can cost around $500,000, and nearly one in 1,000 doses fail due to sterility testing, meaning seriously ill patients will not receive treatment. We realised that an automated system could cut the time taken to find failures – giving patients more opportunity to get their treatment – and reduce the labour-intensive steps in the lab.’
By chance, Gibson and colleagues came across Prophesee’s Metavision event-based sensor around two years ago. They integrated it into an automated contamination detection system that operates in real-time, which ended up as PureSentry.
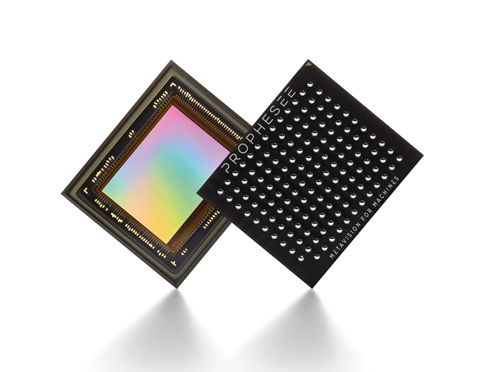
Prophesee’s Metavision event-based sensor. Credit: Prophesee
Inspired by the human retina, Prophesee’s event-based Metavision sensor comprises 300,000 independent and asynchronous pixels that are responsive to low contrast, transient events. These pixels are essentially relative-change detectors, and activate independently according to any change in contrast detected in a scene. When activated, the pixels create a stream of time-stamped events in which their location within the sensor and the direction of the change in brightness - event-on or event-off - are encoded.
‘We have software algorithms that mimic the way the brain leverages visual information... Each intelligent pixel is triggered by motion and decides when to activate,’ explained Guillaume Butin, marketing director at Prophesee. ‘One pixel activating is an event, and we only see what moves.’
What makes event-based sensing different is that an output is only generated when the change in contrast exceeds a threshold. In comparison, conventional cameras will sample every pixel at a fixed rate. Such an event-based approach reduces power, latency and data processing requirements compared to frame-based systems, while achieving much higher dynamic ranges. Events can be recorded that would require conventional cameras to operate at 10,000 images per second or more. And critically, this approach makes label-free imaging a reality, even for low contrast targets, such as cells – spelling good news for cell and microbe monitoring.
‘When we’re flowing our cell culture medium past the microscope, we could be seeing only one bacteria among many millions of human T cells in our millilitre sample,’ said Gibson. ‘So what we really want to do is cut out all of the background and unnecessary data in a scene, and pick out what is important – event-based sensing is great for this as it only responds to fluctuations in brightness.’
As part of the PureSentry closed-loop system, an inverted microscope equipped with Prophesee’s event-based camera captures data on cells as these and associated media are continuously pumped from a bioreactor through a microfluidic cell. Cell data is then sent to post-processing software including an artificial intelligence-event classifier algorithm and decision algorithm to determine if the sample is sterile or contaminated.
‘The event-based sensor simply slotted into existing microscope setups – we could easily compare how this worked compared to standard cameras, and while doing this were able to develop our microfluidic rig,’ said Gibson.
As the physicist pointed out, thanks to the large dynamic range of the sensor, the PureSentry system can use an LED light source rather than laser light, cutting overall costs and reducing the risk of photodamage to cells. The automation software for the system’s hardware was based on Prophesee’s Python APIs, while the Cambridge Consultants researchers developed post-processing software using machine learning recurrent neural network algorithms, which, as Gibson said, work well in an event-based sensing set-up.
‘We can see the cells flowing past the microscope and are only working with the data that we care about,’ he said. ‘[This means] the machine learning algorithms can more easily recognise if those cells are, say, T cells or something more unusual as we don’t have any other changes in the scene.’
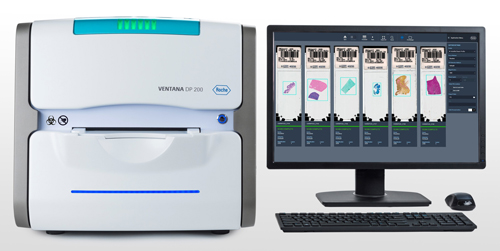
Roche's Ventana 200 promises reliable, high-speed scanning of histology slides for digital pathology. Credit: Roche
So far, Cambridge Consultants researchers have used PureSentry to distinguish between E. coli and T cells in real-time, but the system – with its resolution of around half a micron – could be applied to other contaminants including Staphylococcus aureus and Candida albicans. ‘We’ve distinguished between E. coli and T cells based on size, but we’ve also seen these bacteria rotate and cartwheel as they flow through the system and should be able to distinguish contaminants based on how they move as well as shape,’ said Gibson. ‘We can also monitor the population of different-sized particles over time.’
Gibson is also adamant that PureSentry, with the Prophesee sensor, is set to make a big impact on contamination detection for cell and gene therapy monitoring, as well as other sectors. ‘We could look at yeast cells and contaminants flowing through beer or lactose, and contaminants flowing through milk,’ he said. ‘The prototype is in our lab and we’re looking for a commercial partner to take this forward with us.’
Researchers at France-based CEA-Leti have also developed a novel method that so far targets two key applications. In a similar vein to the PureSentry system, the setup can identify and discriminate between different species of bacteria. However, it can also bring down the time taken to detect cancer in tissue samples from days to minutes.
As scientific director, Dr Laurent Duraffourg, and research engineer, Dr Mathieu Dupoy, noted, for tumour detection, their label-free mid-infrared multispectral method can image a 1cm² tissue sample in minutes using four wavelengths. In contrast, the same sample would take several hours to image using FTIR spectroscopy, and around two days using today’s tumour-biopsy staining and immuno-histochemistry labelling procedures, which demand human assessment to confirm disease.
‘This is a new paradigm for laboratories,’ said Dupoy. ‘We are removing the sample prep and shortening the analysis time – we are providing widefield clinical imaging that is so much faster than classical staining and labelling.’
Their multispectral method comprises an array of mid-infrared quantum cascade lasers, in the 5µm to 11µm wavelength range, and lensless imaging, with an uncooled bolometer matrix, comprising an array of heat-detecting sensors that are sensitive to infrared radiation wavelengths. The setup delivers biochemical mapping over a 2.73 by 2.73mm²-wide field of view in a 20ms measurement time per wavelength.
The laser beams are directed at the tissue, with the infrared radiation transmitted from the tissue then detected by the bolometer matrix. As the infrared radiation is absorbed by a bolometer element, that element increases in temperature, changing its electrical resistance – this change is measured and processed into temperature values that are used to create a mid-infrared absorption image. Such images correspond to the vibrations of targeted chemical bonds, giving a biochemical fingerprint.
According to Dupoy, many of these images can then be combined to create a false-colour image, ready for classification and cancer detection. ‘From these images, you can see if the tissue is healthy or not and you can also get information on the spread of the cancer, if that tissue is cancerous,’ he said.
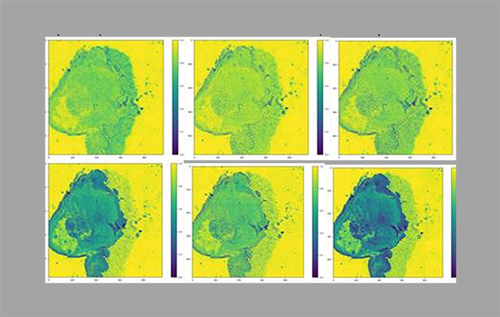
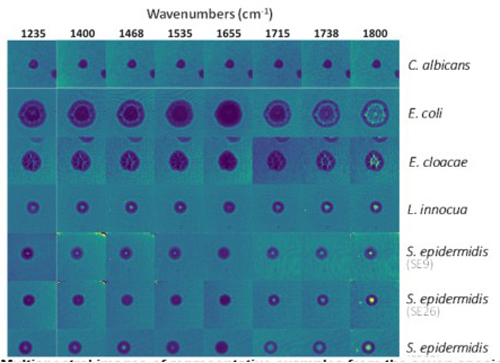
Multispectral lensless imaging from CEA-Leti, used to differentiate tumour cells (top) and bacteria (bottom). Credit: CEA-Leti
Perhaps not surprisingly, the experimental setup is coupled with machine learning algorithms to assist biological cell or bacteria classification in a fast and reproducible way. Indeed, the CEA-Leti researchers have developed machine learning algorithms to work with head and neck cancers, breast cancer, as well as microbial detection. And in recent tumour analyses on mouse tissue, which used 6µm and 10µm infrared wavelengths that are absorbed by endogenous markers (proteins and DNA), the method detected 94 per cent of cancer cells. Likewise, trials on 1,050 colonies of different Staphylococcus strains correctly identified 93 to 96 per cent of the species.
Given the results so far, both Dupoy and Duraffourg are convinced their method, in time, will be used in the digital pathology workflows of tomorrow. Neither CEA-Leti researcher knows of a lab that routinely uses infrared-based imaging in cancer tissue assessment, and are about to complete the development of a portable multispectral lensless imaging setup that can be easily transported to hospitals. Duraffourg is also heading up the launch of a start-up to commercialise the technology.
‘For now, tissues are analysed in our laboratory, but before translating this into a commercial product, we are building a robust, portable demonstrator that we will take to several hospital labs across Europe that we are partnering with,’ said Duraffourg.
Perhaps predictably, the researchers say widespread use of their technology in clinical settings is several years away. Still, their thoughts on industry demand surely echo those across so many life science sectors. As Duraffourg put it: ‘If you talk to clinicians that are also researchers, they really want to have new methods as they are good for science.’


