Recent months have seen significant investment in neuroscience projects, both on this side of the Atlantic and in the US. In September the Human Brain Project, an EU-funded ICT initiative that seeks to advance neuroscience, medicine and computing, announced €89 million in funding for two years’ research. Just one month later the US’s National Institute of Health announced $70 million in funding for projects that will ‘develop new tools and technologies to understand neural circuit function and capture a dynamic view of the brain in action.’
The use of imaging technologies to understand how the brain works has a long and distinguished history. Nearly 40 years ago, Professor Frans Jöbsis of Duke University, North Carolina, became one of the science’s early pioneers when he published a paper entitled ‘Non-invasive, infrared monitoring of cerebral and myocardial oxygen sufficiency and circulatory parameters’ in the journal Science. The paper demonstrated that it was possible to measure blood and tissue oxygenation changes in the brain – in this case of a cat – using near-infrared light.
Since then, the imaging technology that underpins and enables this sector of neurobiology has advanced, with more than 60 papers due to be presented at the Neural Imaging, Sensing, Optogenetics, and Optical Manipulation segment of Photonics West, in San Francisco from 28 January to 2 February 2017.
Both in vivo and in vitro techniques exist to measure neural activity, with gamma ray radiation (PET scans), magnetic fields (MRI scanners) and light (more traditional imaging cameras) used. Each has its merits but, as Elizabeth Hillman, associate professor of biomedical engineering at Columbia University, New York and specialist in advanced in vivo optical imaging, wrote in a 2008 paper: ‘There are significant benefits to using light to image living tissue. Most importantly, light can provide exquisite sensitivity to functional changes, either via intrinsic changes in absorption, fluorescence, or scatter, or through the use of extrinsic contrast.’
Complications of lighting
A typical experiment might look to monitor cell functions under certain stimuli, using genetic techniques to knock out genes, and therefore the proteins or enzymes they encode, to better understand their role in a cell or metabolic pathway. To monitor these typically very fast cellular reactions, fluorescent markers can be tagged onto the gene product (or an antibody that binds with it) which can then be detected by the camera.
The system used to capture this process is relatively simple – a camera and microscope, with an LED or laser light, emitting specific frequencies to activate the fluorescent marker. Alternatively, for mouse in vivo models, a small hole is drilled into the skull with the camera set up against the sedated animal. There are, however, significant complexities to both methods, one of the biggest being lighting. Many neurons analysed – and cells in general – are sensitive to light exposure, a problem called photobleaching. So, there is a trade-off in the amount of light needed to detect the cellular process as it happens and the shortened cell life span that reduces the chance of witnessing this.
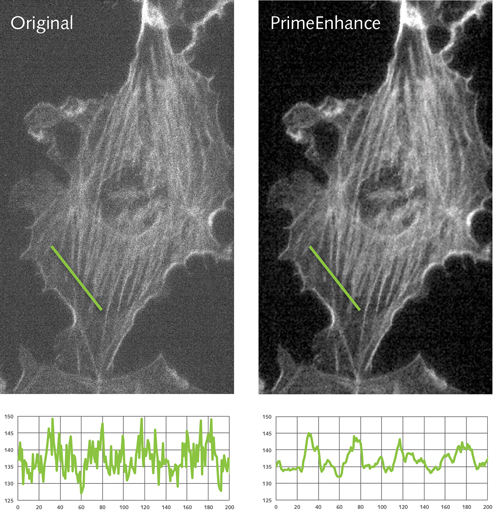
Comparison of a neuron image pre and post de-noising algorithms.
Rachit Mohindra of Photometrics, a developer and supplier of cameras to the life sciences sector, commented that this trade-off can have a large impact depending on what is being imaged and how long a neurological process lasts.
Interesting work to visualise processes in the brain and neurons is also taking place in humans. Teledyne Dalsa has been working closely with Dr Kostadinka Bizheva of the Centre for Bioengineering and Biotechnology at the University of Waterloo in the US. The project has adapted non-invasive optical coherence tomography (OCT) machines, typically used to detect diseases such as glaucoma, with TDI line scan cameras that are able to detect in the near infrared (NIR). The system can generate sub-cellular images that penetrate to a depth of several millimetres within tissue, which then helps clinicians diagnose disease, including brain tumours.
Speaking at a November 2015 conference, Dr Bizheva said of the system: ‘Typically in neurosurgery you don’t have the time to do a biopsy and then do the normal stained histology. With our high-res system we can recognise the large nuclei of cancer cells, and we can differentiate between healthy grey matter and different types of tumour.’
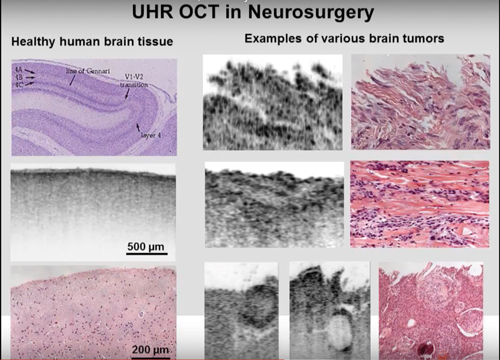
OCT Imaging can identify and distinguish between multiple tumour classes, as presented by Dr Bizheva in November 2015 at the University of Waterloo
Teledyne Dalsa’s Geralyn Miller commented: ‘As a fairly new medical practice, UHR-OCT has a long way to go before we will realise its full potential. As the imaging technology advances so will OCT more generally, ultimately helping to broaden our understanding of the human body.’
Sensor technology
Advances in sensor technology have the potential to significantly improve detection. For neural imaging the industry has traditionally had a choice of two major classes of sensor: electron multiplication CCD (EMCCD) or scientific CMOS (sCMOS). Cameras based on these have been available from a wide range of companies such as Andor, Alrad, PCO, Photometrics and QImaging. Photometrics’ Mohindra stated: ‘EMCCD cameras have the ability to multiply and amplify a signal above any noise detection limits that are in place. This allows you to detect these extremely dim signals and see them very quickly.’
Mohindra explained that it’s challenging to capture fast biological processes because of the lack of light. ‘This is what EMCCDs very much excelled at, because they [offer] almost perfect levels of sensitivity and have the frame rates that allow these high rates of capture.’
Conversely, Mohindra said: ‘Scientific CMOS … allows you to have a very large sensor that is very sensitive and goes very fast. With CCDs you had to take two of the performance factors and the other was the trade-off – for example if I had a large sensor that typically meant I was going to have a large number of megapixels… or if I needed sensitivity, then typically resolution is what I’d be trading off.’
Scientific CMOS means researchers can capture four megapixel images at 100 frames per second, Mohindra said, and with smaller regions of interest to go ‘well into the thousands of frames per second’.
However, there were still trade-offs on sensitivity. Mohindra said: ‘The EMCCD’s conversion factor [quantum efficiency] of measured signal is about 95 per cent. That’s about as good as you’re going to get. The sCMOS cameras were – when they first came out – [about] 60 per cent. Over the years this improved to about 70 or 80 per cent; this was getting close to where EMCCD cameras were, but every bit of sensitivity helps.’

The Prime 95b camera using Photometrics' GSense sCMOS sensor
In June Photometrics launched the GSense sCMOS sensor, already developed into its Prime 95B camera using the PCIe transmission standard. This sensor, the company claims, delivers a QE that matches EMCCD’s 95 per cent. The 3/4-inch sensor delivers 1.44 megapixel (1,200 x 1,200 pixels) images with a median read noise of 1.3 electrons (1.5rms). The sensor can detect just two photons hitting the sensor; it has a combined gain of 80,000e-, which Mohindra claims is approximately 2.5 times that of the next best sCMOS sensor.
The sensor achieves this level of sensitivity through a backside illumination process, moving the lenses and electrical structures that would typically be in the way, so the light hits the sensitive area first, and the majority of it is converted to charge.
Mohindra said that this process of back thinning ‘isn’t a novel idea... the ability to do it on a CMOS device with large pixels has been a challenge for quite a long time,’ adding ‘there are [a lot] of experimental factors that need to be balanced in the imaging. And that’s where the sensitivity really helps, so if you’re able to maximise your light collection, you are able to turn down the intensity of the light you’re using to photograph your sample and keep your cells alive for longer.’
Future developments
Assuming the GSense sCMOS sensor lives up to its hype – and it’s already in use at the Berkley campus of the University of California and the Salk Institute – then scientists and medics can now detect about a photon’s worth of signal, allowing almost any cellular process to be monitored. Couple this with exceptionally high frame rates – into the thousands per second – and the advances become less about the industry’s standard incremental improvements of megapixels and frame rates. So how will these cameras evolve?
Mohindra said the industry needs to ‘provide these answers easier or faster’. Scientific cameras are already following trends adopted by the wider imaging sector, developing smart cameras that bring in FPGAs and enable real-time modification of image data. Mohindra noted: ‘Typically with imaging the first challenge is “Can I see it?” and the second, “How well can I see it?”
‘For example, one of the algorithms we’ve put onto the camera is a de-noising algorithm. This allows people to get, in real time, images that look clearer with less variation and less uncertainty in them. This enables you to reduce the level of light or excitation of the cell to get the same quality of data, [which] directly translates into keeping cells alive for longer. So your experiments become easier to run for longer amounts of time.’
Another challenge is that of handling the amount of data produced by these sensors – a four megapixel camera with 100 frames per second generates roughly 800MB/s, according to Mohindra.
The scientific camera sector is implementing features – such as region of interest – seen in the wider vision sector. However, advances in scientific imaging will also come from new biological techniques. Professor Takeharu Nagai at Osaka University in Japan has developed a ‘Nano-Lantern’ probe, which fuses a luminescent protein from the sea pansy (Renilla reniformis) with a previously designed fluorescent protein. This gives the brightest luminescence of any known protein – it generates its own light without having to be activated by a light source – and has a temporal and spatial resolution equivalent to fluorescence. According to Professor Nagai: ‘Dispensing with the need for light illumination – which is essential for fluorescence imaging – allows us to analyse dynamics in environments where the use of fluorescent indicators is not feasible.’
With significant funding from the US and European governments boosting neuroscience, the vision sector will continue to work closely with research institutes to develop the technology in line with the experiments.